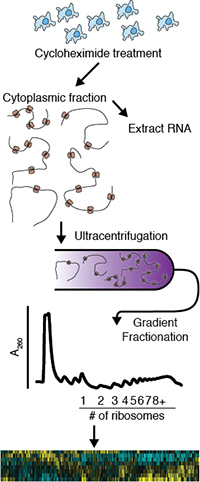
Eukaryotic genes generate multiple mRNA transcript isoforms though alternative transcription, splicing, and polyadenylation. However, the relationship between human transcript diversity and protein production is complex as each isoform can be translated differently. To understand this interplay, we fractionated a polysome profile and reconstructed transcript isoforms from each fraction, which we term Transcript Isoforms in Polysomes sequencing (TrIP-seq). Analysis of these data revealed regulatory features that control ribosome occupancy and translational output of each transcript isoform. We extracted a panel of 5′ and 3′ untranslated regions that control protein production from an unrelated gene in cells over a 100-fold range. Select 5′ untranslated regions exert robust translational control between cell lines, while 3′ untranslated regions can confer cell-type-specific expression. These results expose the large dynamic range of transcript-isoform-specific translational control, identify isoform-specific sequences that control protein output in human cells, and demonstrate that transcript isoform diversity must be considered when relating RNA and protein levels. Read more…
Proper maintenance of RNA structure and dynamics is essential to maintain cellular health. Multiple families of RNA chaperones exist in cells to modulate RNA structure, RNA–protein complexes, and RNA granules. The largest of these families is the DEAD-box proteins. The human DEAD-box protein DDX3 is implicated in diverse biological processes including translation initiation and is mutated in numerous cancers. Like many DEAD-box proteins, DDX3 is essential to cellular health. Therefore, chemical inhibition would be an ideal tool to probe the function of DDX3. However, most DEAD-box protein active sites are extremely similar, complicating the design of specific inhibitors. We found that a chemical genetic approach best characterized in protein kinases, known as analog-sensitive chemical inhibition, is viable for DDX3 and possibly other DEAD-box proteins. We discovered an expanded active-site mutant that is tolerated in vitro and in vivo, and is sensitive to chemical inhibition by a novel bulky inhibitor. Our results highlight a course towards analog sensitive chemical inhibition of DDX3 and potentially the entire DEAD-box protein family. Read more…