Center for Genomic Editing and Recording (CGER), one of the Centers for Excellence in Genomic Science (CEGS)
Research
The Center for Genomic Editing and Recording (CGER) is an NIH-funded collaboration between the Doudna, Joung, Liu, and Weissman labs that is seeking to build on CRISPR-Cas9 genome engineering technology to expand greatly our ability to detect, alter and record the sequence and output of the genome in individual cells and tissues.
Aim I. Develop next-generation genome editing technologies to enable rapid, efficient and accurate genome alterations. We will create methods to generate and detect the broadest possible set of desired sequence alterations in virtually any locus with high efficiency and specificity.
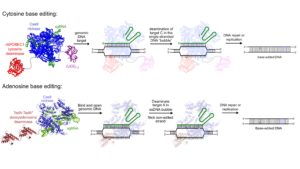
Aim II. Implement a high-throughput pipeline to identify and interrogate suppressors of sequence variants associated with human disease. We will identify and illuminate the relationships between human sequence variation, gene expression and interactions among groups of genes and regulatory elements. This will be enabled by the development of new methods, based on single cell RNA-seq, for the massively parallel monitoring of changes in the mRNA output of cells in response to perturbations in gene expression.
Aim III. Create robust cell data recording technologies with single-cell RNA seq readout. We will develop new methods for permanently recording cell state changes in DNA so that they can be read out in a single-cell format. We will focus on building, validating and using a Cas9-based evolvin
g lineage tracer that expresses these DNA barcodes as mRNAs. Paired with droplet-based single cell RNA-seq technology, this will permit us to trace the lineage of hundreds of thousands of cells along with their functional cell state as revealed by their RNA-seq signature, thus connecting the cell’s present state with its past history. We will produce a toolkit for molecular recorders, including the biological reagents required for their execution and the computational framework needed for their analysis. Finally, we will extend our molecular recorder approach to capture other cellular information such as the timing of differentiation, signaling activity and acquisition of mutations.
Executive Committee

Dr. Jennifer Doudna is a member of the departments of Molecular and Cell Biology and Chemistry at UC Berkeley, the Howard Hughes Medical Institute, and Lawrence Berkeley National Lab, along with the National Academy of Sciences, and the American Academy of Arts and Sciences.
Biographical Highlights:
- Fellow, American Academy of Arts and Sciences (2003)
- Professor of Biochemistry and Molecular Biology, Department of Molecular and Cell Biology, the University of California, Berkeley (2003)
- Professor of Biochemistry and Molecular Biology, Department of Chemistry, the University of California, Berkeley (2003)
- Faculty, Biophysics Graduate Group, the University of California, Berkeley (2003)
- Faculty Scientist, Physical Biosciences Division, Lawerence Berkeley National Laboratory (2003)
- Member, National Academy of Sciences (2002)
- Member, Board of Trustees, Pomona College (2001)
- American Chemical Society Eli Lilly Award in Biological Chemistry (2001)
- R. B. Woodward Visiting Professor, Harvard University (2000-2001)
- Alan T. Waterman Award (2000)
- Investigator, Howard Hughes Medical Institute (1997)
- Searle Scholar, Kinship Foundation’s Searle Scholars Program (1996)
- Henry Ford II Professor of Molecular Biophysics and Biochemistry, Center for Structural Biology, Department of Molecular Biophysics and Biochemistry, Yale University (1994-2002)
- Lucille P. Markey Scholar in Biomedical Science, University of Colorado (1991-1994, Dr. Thomas R. Cech)
- Postdoctoral Research Fellow, Molecular Biology, Massachusetts General Hospital and Harvard Medical School (1989-1991, Dr. Jack W. Szostak)
- Ph.D. Harvard University (1989, Dr. Jack W. Szostak)
- B.A. Pomona College (1985, Dr. Sharon M. Panasenko)
Feel free to use this short bio and picture for announcements.

J. Keith Joung is currently Desmond and Ann Heathwood Research Scholar, Pathologist, and Associate Chief of Pathology for Research at Massachusetts General Hospital (MGH) and is Professor of Pathology at Harvard Medical School. He is also a member of the Center for Cancer Research and the Center for Computational and Integrative Biology at MGH. Dr. Joung has been a pioneer in the development of important technologies for targeted genome editing and epigenome editing of human cells. He has received numerous awards including an NIH Director’s Pioneer Award, an NIH Director’s Transformative Research Project R01 Award, the MGH Research Scholar Award, an NIH R35 MIRA (Maximizing Investigators Research Award), election into the American Association of University Pathologists, and designation as a “Highly Cited Researcher” in 2016 and 2017 by Thomson Reuters/Clarivate Analytics. He serves on the Board of Directors for the American Society of Gene and Cell Therapy and the editorial boards of Genome Biology, Human Gene Therapy, and Trends in Biotechnology. Dr. Joung holds a Ph.D. in genetics from Harvard University, an M.D. from Harvard Medical School and an A.B. in biochemical sciences from Harvard College.

David R. Liu is the Richard Merkin Professor, Director of the Merkin Institute of Transformative Technologies in Healthcare, and Vice-Chair of the Faculty at the Broad Institute of Harvard and MIT; Professor of Chemistry and Chemical Biology at Harvard University; and Howard Hughes Medical Institute Investigator. Liu graduated first in his class at Harvard in 1994. He performed synthetic organic and bioorganic chemistry research on sterol biosynthesis under Professor E. J. Corey’s guidance as an undergraduate. During his Ph.D. research with Professor Peter Schultz at U. C. Berkeley, Liu initiated the first general effort to expand the genetic code in living cells. He earned his Ph.D. in 1999 and became Assistant Professor of Chemistry and Chemical Biology at Harvard University in the same year. He was promoted to Associate Professor in 2003 and to Full Professor in 2005. Liu became a Howard Hughes Medical Institute Investigator in 2005 and joined the JASONs, academic science advisors to the U.S. government, in 2009. Liu has earned several university-wide distinctions for teaching at Harvard, including the Joseph R. Levenson Memorial Teaching Prize, the Roslyn Abramson Award, and a Harvard College Professorship. He has published more than 150 papers and is the inventor of more than 50 issued patents. His research accomplishments have earned distinctions including the Ronald Breslow Award for Biomimetic Chemistry, the American Chemical Society Pure Chemistry Award, the Arthur C. Cope Young Scholar Award, and awards from the Sloan Foundation, Beckman Foundation, NSF CAREER Program, and Searle Scholars Program. In 2016 he was named one of the Top 20 Translational Researchers in the world by Nature Biotechnology, and in 2017 was named to the Nature’s 10 researchers in world and to the Foreign Policy Leading Global Thinkers. Professor Liu’s research integrates chemistry and evolution to illuminate biology and enable next-generation therapeutics. His major research interests include the engineering, evolution, and in vivo delivery of genome editing proteins such as base editors to study and treat genetic diseases; the evolution of proteins with novel therapeutic potential using phage-assisted continuous evolution (PACE); and the discovery of bioactive synthetic small molecules and synthetic polymers through DNA-templated organic synthesis. Base editing (named one of four 2017 Breakthrough of the Year finalists by Science), PACE, and DNA-templated synthesis are three examples of technologies pioneered in his laboratory. He is the scientific founder or co-founder of several biotechnology and therapeutics companies, including Ensemble Therapeutics, Permeon Biologics, Editas Medicine, Pairwise Plants, and Beam Therapeutics.

Jonathan Weissman, Ph.D., studies how cells ensure that proteins fold into their correct shape, as well as the role of protein misfolding in disease and normal physiology. He is also widely recognized for building innovative tools for broadly exploring organizational principles of biological systems. These include ribosome profiling, which globally monitors protein translation, and CRIPSRi/a for controlling the expression of human genes and rewiring the epigenome. Dr. Weissman is a professor at the University of California San Francisco and an Investigator at the Howard Hughes Medical Institute. He is a member of the National Academy of Sciences, a member of the Scientific Advisory Board for Amgen, co-director of the Innovative Genome Initiative of Berkeley and UCSF, and a member of the President’s Advisory Group for the Chan-Zuckerberg Biohub. Dr. Weissman has received numerous awards including the Beverly and Raymond Sackler International Prize in Biophysics (2008), The Keith Porter Award Lecture from the American Society of Cell Biology (2015) and the National Academy Science Award for Scientific Discovery (2015).
New Collaborators
Eric Chow is an Adjunct Assistant Professor in the Department of Biochemistry and Biophysics at the University of California, San Francisco. He earned his PhD under Jeffrey Cox at UCSF studying M. tuberculosis virulence using genomics and lipidomics. Eric also directs the UCSF Center for Advanced Technology (CAT), a core that houses technology and equipment in a central facility accessible to all researchers. The CAT has a major focus on genomics. Eric’s lab focuses on the development of new tools in the NGS space with goals of making methods, cheaper, better, and faster. His lab has a strong interest pushing applications in the single-cell profiling space.

Maria Jasin is biomedical researcher at Memorial Sloan Kettering Cancer Center (MSK) in New York, where she holds the William E. Snee Chair in the Developmental Biology Program. Research in her laboratory focuses on DNA recombination and the relationship to the maintenance of genomic integrity and cancer, targeted genome modification, and meiosis. Discoveries from her laboratory include defining how chromosome double-strand breaks are repaired, accomplishing the first “gene editing” experiment, and determining a role for the breast cancer suppressors BRCA1 and BRCA2 in recombination. She obtained a Ph.D. from the Massachusetts Institute of Technology performing thesis research in course VII the laboratory of Paul Schimmel. Postdoctoral research was performed at the University of Zürich in Walter Schaffner’s laboratory and Stanford University in Paul Berg’s laboratory in the Department of Biochemistry in the Stanford Medical School before starting her own lab in New York in 1990. In addition to her MSK appointment, she also has an appointment in the Weill Cornell Graduate School of Medical Sciences.

Luca is an Assistant Professor at Massachusetts General Hospital and Harvard Medical School. He received his BA,MA and Phd in Computer Science and Mathematics from the University of Palermo in Italy. He had the good fortune to be part of the “omics” revolution and the opportunity to work on many computationally-challenging problems since he was an undergraduate student. During his postdoctoral research at Dana-Farber Cancer Institute/Harvard School of Public Health, he studied the role of chromatin structure in gene regulation and developed computational methods for single cell analysis and epigenomics. He has a background in computer science and extensive experience in machine learning, data mining and web technologies. His research program uses computational approaches to systematically analyze the sources of variation that affect gene regulation: epigenetic variation, genetic variation and (single-cell) gene expression variability. He is actively involved in the single-cell community and he is part of the Human Cell Atlas initiative, proposing computational strategies to model gene expression variability, its relationship with chromatin accessibility and DNA methylation, and to reconstruct developmental trajectories (http://stream.pinellolab.org). He fully embraced the revolution in functional genomics made possible by the novel genome editing approaches such as CRISPR/Cas9. He developed computational tools to quantify and visualize the outcome of genome editing experiments, that are nowadays the standard the facto for the community (http://crispresso2.pinellolab.org). He also a subgroup coordinator for the NIST Consortium on genome editing. His contributions center on defining standards for data obtained from genome editing experiments. In his free time, he likes to hike, play his guitar and roast green coffee.

David F. Savage is an Associate Professor of Biochemistry, Biophysics, and Structural Biology in the Department of Molecular & Cell Biology at the University of California, Berkeley. As an undergraduate, Dave attended Gustavus Adolphus College, where he majored in Chemistry and minored in Computer Science. He received his Ph.D. in 2007 from UCSF for his work on membrane protein structure determination with Robert Stroud. From 2007 to 2011 Dave was a Life Sciences Research Foundation fellow with Pamela Silver in the Department of Systems Biology at Harvard Medical School. At Berkeley, his lab focuses on developing new tools for the understanding and engineering of biology with an emphasis on the protein machineries associated with genome editing and CO2 assimilation.