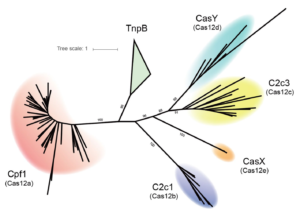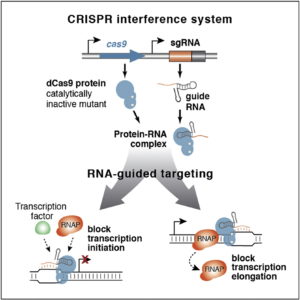
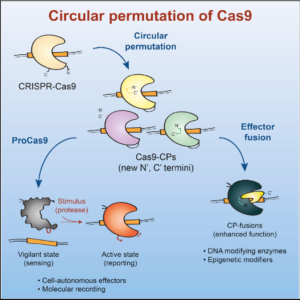
The Doudna lab employs protein engineering and screening strategies to bestow CRISPR-Cas enzymes with improved and novel functionalities. In collaboration with Stanley Qi and the laboratories of Wendell Lim, Jonathan Weissman, and Adam Arkin, we pioneered the engineering of CRISPR-Cas proteins for new purposes by converting Cas9 into a tool for targeted control of gene expression, referred to as CRISPR interference (CRISPRi). In this platform, catalytically inactive Cas9 serves as an RNA-guided DNA binding protein to silence gene expression in a sequence-specific manner. In collaboration with the laboratory of David Savage, we have also developed a series of high-throughput mutant library construction, screening, and sequencing pipelines to probe CRISPR-Cas protein function and to isolate engineered variants with novel properties. In one example, we utilized domain insertion profiling with sequencing (DIP-seq) to identify an allosteric Cas9 switch, yielding an optimized Cas9 variant that is activated by the small molecule 4-hydroxytamoxifen. Recently, we used a similar large-scale protein engineering and screening platform to construct circularly permuted, viral protease-activated ProCas9s that orchestrate programmed cellular responses to pathogen-associated protease activity.