Structure and Mechanism
DNA Acquisition in CRISPR-Cas systems
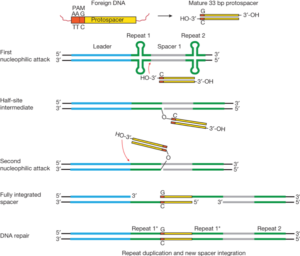 CRISPR-harboring organisms generate immunological memory of previous infections by capturing short segments of foreign DNA for integration into CRISPR loci as spacer sequences. Central to this process are Cas1 and Cas2 – which are generally conserved in most CRISPR systems. Using a combination of biochemical, structural, and genetic approaches, we found that Cas1 and Cas2 functions as a protein complex. The E.coli Cas1–Cas2 complex (Type IE) captures ~30 bp of foreign DNA and integrates them into the CRISPR locus via a direct nucleophilic reaction similar to many retroviral integrases and transposases. Our results uncover the structural basis for foreign DNA capture and the mechanism by which Cas1–Cas2 functions as a molecular ruler to dictate the sequence architecture of CRISPR loci. We have also demonstrated how an integration host factor helps Cas1-Cas2 recognize the CRISPR array for site-specific integration. Recently, we found that a mini-integrase comprising the type V-C Cas1 protein alone catalyzes DNA integration with a shortened ruler mechanism, which may give insight to the function of the ancestral Cas1, prior to Cas2 adoption.
CRISPR-harboring organisms generate immunological memory of previous infections by capturing short segments of foreign DNA for integration into CRISPR loci as spacer sequences. Central to this process are Cas1 and Cas2 – which are generally conserved in most CRISPR systems. Using a combination of biochemical, structural, and genetic approaches, we found that Cas1 and Cas2 functions as a protein complex. The E.coli Cas1–Cas2 complex (Type IE) captures ~30 bp of foreign DNA and integrates them into the CRISPR locus via a direct nucleophilic reaction similar to many retroviral integrases and transposases. Our results uncover the structural basis for foreign DNA capture and the mechanism by which Cas1–Cas2 functions as a molecular ruler to dictate the sequence architecture of CRISPR loci. We have also demonstrated how an integration host factor helps Cas1-Cas2 recognize the CRISPR array for site-specific integration. Recently, we found that a mini-integrase comprising the type V-C Cas1 protein alone catalyzes DNA integration with a shortened ruler mechanism, which may give insight to the function of the ancestral Cas1, prior to Cas2 adoption.
Figure legend: Integration occurs via nucleophilic attacks by the 3’ OH of the protospacer on either side of the leader-adjacent repeat. Link: https://www.nature.com/articles/nature14237
Nucleic Acid Targeting CRISPR-Cas Systems
We use biochemical activity assays in conjunction with structural techniques and sequencing to investigate RNA-targeting CRISPR systems, such as the Type VI effector, Cas13a. A crystal structure of L. bacterium Cas13a bound to a single guide RNA (sgRNA) demonstrated, together with biochemical studies, how crRNAs are processed by this enzyme and how catalytic activity is blocked before target binding. (Knott et al., 2017) We also use high-throughput sequencing, together with biochemistry, to discover mechanisms governing RNA-targeting CRISPR effectors, such as the decoupled RNA targeting and HEPN nuclease activity in LbaCas13a. (Tambe et al, 2018)
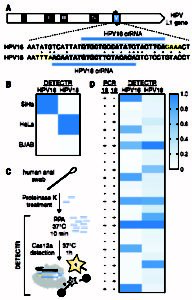 RNA-Targeting CRISPR-Cas Systems – We use biochemical activity assays in conjunction with structural techniques and sequencing to investigate RNA-targeting CRISPR systems, such as the Type VI effector, Cas13a. A crystal structure of L. bacterium Cas13a bound to a single guide RNA (sgRNA) demonstrated, together with biochemical studies, how crRNAs are processed by this enzyme and how catalytic activity is blocked before target binding. (Knott et al., 2017) We also use high-throughput sequencing, together with biochemistry, to discover mechanisms governing RNA-targeting CRISPR effectors, such as the decoupled RNA targeting and HEPN nuclease activity in LbaCas13a. (Tambe et al, 2018)
RNA-Targeting CRISPR-Cas Systems – We use biochemical activity assays in conjunction with structural techniques and sequencing to investigate RNA-targeting CRISPR systems, such as the Type VI effector, Cas13a. A crystal structure of L. bacterium Cas13a bound to a single guide RNA (sgRNA) demonstrated, together with biochemical studies, how crRNAs are processed by this enzyme and how catalytic activity is blocked before target binding. (Knott et al., 2017) We also use high-throughput sequencing, together with biochemistry, to discover mechanisms governing RNA-targeting CRISPR effectors, such as the decoupled RNA targeting and HEPN nuclease activity in LbaCas13a. (Tambe et al, 2018)
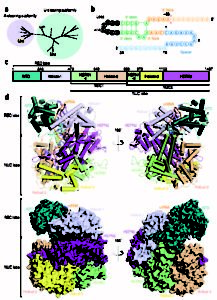 DNA-Targeting CRISPR-Cas Systems – Biochemistry allows us to investigate the mechanisms driving DNA-targeting CRISPR effectors on a molecular level. These studies uncovered indiscriminate, single-stranded DNAase activity in Cas12a unleashed by target binding, the chemistry behind the DETECTR technology (vide infra). (Chen et al., 2018) Structural (Cryo-EM), biochemical, and live-cell studies, as part of a collaboration with the laboratory of Eva Nogales, moreover characterized the fundamental properties of distinct, DNA-targeting CRISPR-CasX effectors. (Liu, Orlova, Oakes, et al., 2019) These studies, in combination with techniques such as high-throughput sequencing and live-cell imaging, provide important insight that helps better engineer and control CRISPR genome editing tools.
DNA-Targeting CRISPR-Cas Systems – Biochemistry allows us to investigate the mechanisms driving DNA-targeting CRISPR effectors on a molecular level. These studies uncovered indiscriminate, single-stranded DNAase activity in Cas12a unleashed by target binding, the chemistry behind the DETECTR technology (vide infra). (Chen et al., 2018) Structural (Cryo-EM), biochemical, and live-cell studies, as part of a collaboration with the laboratory of Eva Nogales, moreover characterized the fundamental properties of distinct, DNA-targeting CRISPR-CasX effectors. (Liu, Orlova, Oakes, et al., 2019) These studies, in combination with techniques such as high-throughput sequencing and live-cell imaging, provide important insight that helps better engineer and control CRISPR genome editing tools.
-
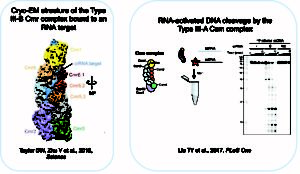 Dual RNA and DNA targeting CRISPR-Cas systems – We use a combination of biochemical and structural biology techniques (single-particle EM) to investigate the mechanism and function of dual DNA- and RNA-targeting Type III CRISPR-Cas systems. In collaboration with the laboratories of Eva Nogales, Akeo Shinkai, and John van der Oost, we have revealed the molecular architecture and RNA targeting mechanism of T. thermophilus Type III-B CRISPR complexes. In collaboration with Eva Nogales’ lab, we are also investigating the pathway of transcription-coupled DNA and RNA targeting by Type III-A CRISPR-Cas systems. Insights into how Type III interference complexes recognize and degrade their targets may lead to improved tools for controlling gene expression in vivo.
Dual RNA and DNA targeting CRISPR-Cas systems – We use a combination of biochemical and structural biology techniques (single-particle EM) to investigate the mechanism and function of dual DNA- and RNA-targeting Type III CRISPR-Cas systems. In collaboration with the laboratories of Eva Nogales, Akeo Shinkai, and John van der Oost, we have revealed the molecular architecture and RNA targeting mechanism of T. thermophilus Type III-B CRISPR complexes. In collaboration with Eva Nogales’ lab, we are also investigating the pathway of transcription-coupled DNA and RNA targeting by Type III-A CRISPR-Cas systems. Insights into how Type III interference complexes recognize and degrade their targets may lead to improved tools for controlling gene expression in vivo.



